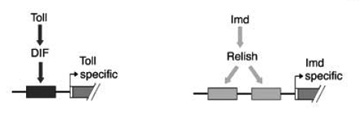
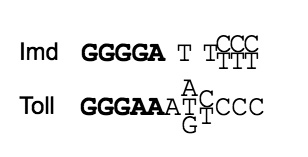Two systems of pathogen recognition and signal transduction mediate the humoral immune response in Drosophila melanogaster. The Imd pathway mediates responses to Gram (-) bacteria and Gram (+) bacilli; the Toll pathway, mediates responses to fungi and other Gram (+) bacteria. Both pathways activate transcription factors in the NF-kappaB/Rel family.
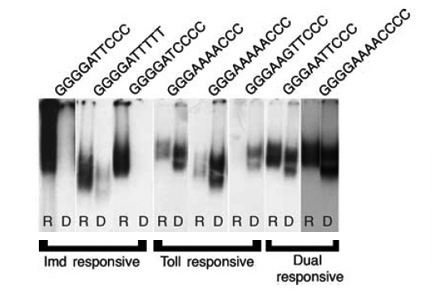
Using genetic, bioinformatic, and biochemical approaches, we have elucidated the sequence code that enables elaboration of specific effector responses. Combining expression data from wild-type and mutant backgrounds with an unbiased analysis of upstream regulatory regions, we defined consensus kappaB sites responsive to each pathway (1).
Recent Research
We expanded our analysis of transcriptional responses to innate immune signaling pathways across a range of Drosophila species, focusing initially on Toll signaling. These studies revealed strong conservation of the sequence code for Toll responsiveness. Global analysis of effector genes by RNAseq defined a core set of Toll responsive genes shared across the genus, as well as species specific effectors. We used a molecular genetic approach to delineate the function of these effectors, for many of which the innate immune function had not previously been characterized.