Studying evolution is notoriously difficult because it relies on multiple stochastic processes and the predictable process, natural selection, operates through a genotype to phenotype to fitness map that is riddled with nonadditive interactions like epistasis. An even more difficult challenge is studying coevolution because the evolution of multiple species has to be accounted for, and their evolution depends on one another creating a tangled bank of high order biological interactions. To untangle the bank, we have developed two multispecies laboratory systems that provide experimental control required to build a mechanistic understanding of coevolution.
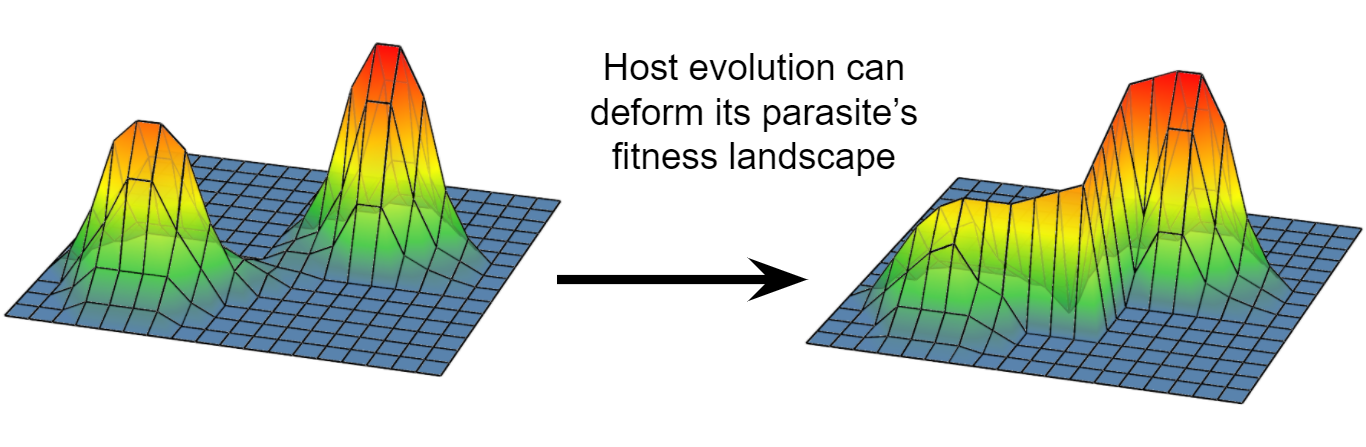
We have published many papers on coevolution between bacteriophage lambda and its host, Escherichia coli. We are currently interested in using this model system to study how fitness landscapes are deformed during coevolution and how these deformations can set up evolutionary feedbacks that promote innovation.
Microbiome research has proliferated in the last decade. Our role in this area has been to develop a model experimental system to study interactions between metazoan hosts and their microbiomes. The host is a rotifer that has a number of characteristics amenable to microbiome evolution experiments; such as rapid generation time, ease of culturing large population sizes, and it has a rudimentary gut microbiome. We developed this system to learn how hosts coevolve with their microbiomes and to determine how microbiome effects extend beyond their host and into the broader ecosystem.