Regulation of Human mRNA Turnover
mRNA turnover plays a critical role in regulation of gene expression. With the long-term goal of understanding how mRNA decay is regulated in gene expression and disease, our lab focuses on dissecting the human cellular mRNA decay machineries.
mRNA Decay in Human Cells
Correct regulation of gene expression is essential for the growth and development of an organism. The expression of genes in the cell is to a large extent controlled at the level of mRNA accumulation. One way by which the level of a specific mRNA can be regulated is by controlling its rate of decay. The accurate regulation of mRNA turnover is a pre-requisite for sustained life, and mis-regulation of mRNA decay is associated with a range of human diseases. However, despite its importance in gene expression, the mechanism and regulation of human mRNA decay is currently poorly understood.
Many mRNAs encoding proto-oncogenes, interleukins and transcription factors contain destabilizing AU-rich elements (AREs) in their 3' untranslated regions (UTRs). Specific cell signals can transiently stabilize these mRNAs and thereby upregulate protein expression by orders of magnitude. Mis-regulation of ARE-mediated mRNA decay can lead to transformation of human cells, leading to tumor growth.
Another mRNA decay pathway, termed nonsense-mediated decay (NMD), targets mRNAs that have acquired premature translation termination codons due to failure in mRNA processing or to genetic mutation. This pathway is believed to render recessive many human genetic diseases, such as beta-thalasemia, marfans syndrome, cystic fibrosis etc, by degrading mRNAs that originate from the mutant alleles with premature termination codons. NMD also plays an important role in regulating mRNA levels from normal genes.
Eukaryotic mRNAs are protected at their 5' and 3' ends by the cap and poly(A) tail. The limiting step in decay of most mRNAs is the removal of these elements. In Saccharomyces cerevisiae, mRNAs are predominantly degraded by initial removal of the poly(A) tail, by deadenylation, followed by decapping and 5'-3' and 3'-5' exonucleolytic decay. Many of the enzyme complexes involved in these processes have been identified by genetic screens. In contrast, very little is known about human mRNA decay factors.
Current Lab Projects
Our laboratory focuses on several aspects of mRNA decay regulation in human cells, described below.
1) Regulation of ARE-mediated mRNA Decay:
An important signal for rapid mRNA turnover in mammalian cells is the AU-rich element (ARE), which is found in the 3'UTR of many unstable mRNAs. Importantly, while such mRNAs are normally rapidly degraded, specific cell signals can trigger mRNA stabilization and result in protein production. For example, ARE-mediated decay plays a prominent role in the regulation of mRNAs that encode proto-oncogenes, interleukins and cytokines. The ARE-binding protein TTP is an activator of ARE-mediated decay in human cells. TTP contains two CCCH-type zinc finger domains that are necessary and sufficient for ARE binding. In addition, two paralogs of TTP involved in ARE-mediated decay are BRF-1 and BRF-2. To understand how the TTP-family of ARE-binding proteins activate mRNA decay, we are investigating how they communicate with enzymes involved in mRNA decay. This should provide a first step to understanding the mechanism by which ARE-mediated decay can be regulated at the molecular level, to modulate gene expression.
2) The Mechanism of Nonsense-Mediated Decay:
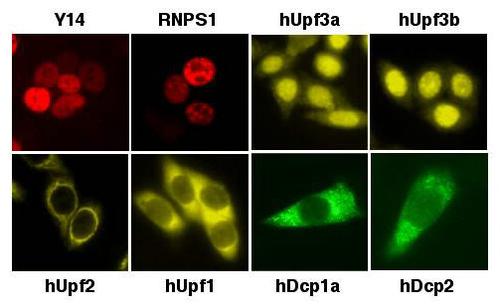
Eukaryotic cells have evolved mechanisms to ensure the fidelity of gene expression. One such mechanism, called mRNA surveillance, ensures that only mRNAs with full coding potential are available for translation in the cytoplasm. This process detects mRNAs with truncated open reading frames and subjects them to nonsense-mediated mRNA decay (NMD). NMD thus prevents the synthesis of potentially deleterious truncated proteins and is responsible for rendering a large fraction of human disease mutations recessive. We, and others, previously identified human proteins involved in NMD, named hUpf1, hUpf2 and hUpf3. NMD depends on active translation, and the translation release factors, eRF1 and eRF3, interact with the hUpf proteins. In mammals, a premature termination codon is detected when present in the mRNA more than 50 nucleotides upstream of the last splice junction. We, and others, previously demonstrated that a multi-protein exon-junction complex (EJC), which is deposited upstream of exon-exon junctions after pre-mRNA splicing, communicates with the terminating ribosome via the hUpf proteins to identify the premature termination codons. We are currently conducting experiments to understand how the EJC communicates with the hUpf proteins and the terminating ribosome.
3) Deadenylation in Human mRNA Decay:
Deadenylation is believed to be an important rate-limiting step in mRNA decay. However, very little is known about deadenylases in human cells. Based on sequence similarity to a yeast deadenylase complex, we have identified ten putative human deadenylases. We are currently trying to establish the role of the deadenylases in the ARE- and nonsense-mediated decay pathways (see above).
4) Decapping in Human mRNA Decay :
Another important process in mRNA decay is decapping. We identified a human decapping complex that contains at least two proteins, hDcp1 and hDcp2, and we showed that the hDcp2 protein possesses catalytic activity (Lykke-Andersen, MCB, 2002). We affinity purified the decapping complex from a human cell line, and identified three new proteins in the complex. We are currently testing the importance of these novel proteins in mRNA decapping and decay.