Efficient in vivo and in vitro guide RNA synthesis using ribozyme-flanked construct for CRISPR systems
Yangbin Gao and Yunde Zhao
University of California San Diego
To overcome the limitations of in vivo and in vitro guide RNA production inherited from RNA polymerase III and phage promoters, we have designed a versatile method to efficiently synthesize guide RNA in vitro and in vivo (Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. DOI: 10.1111/jipb.12152).

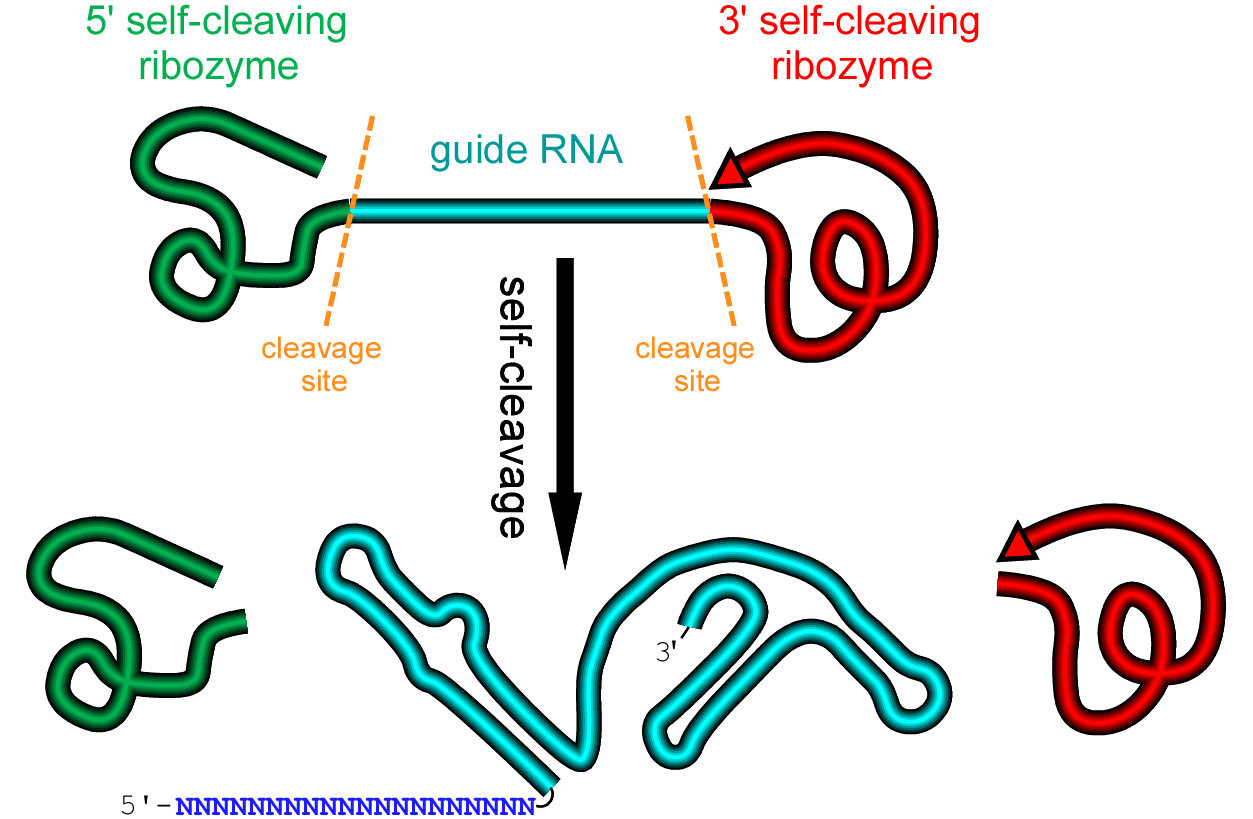
The idea is to attach a self-cleaving ribozyme module onto each end of the guide RNA to generated a ribozyme-gRNA-ribozyme (RGR) cassette. After the RGR cassette is transcribed, the guide RNA will be released from the primary transcript automatically. This removes all the constrains on the target sequence and promoter selection for the guide RNA production, both in vitro and in vivo.
To learn more about how to design a ribozyme-gRNA-ribozyme (RGR)cassette, click here.
For possible applications of our RGR cassette design, click here.
Background
Although a relatively recent discovery, the applications of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) systems have been revolutionizing many fields in biological research, from precise genome editing to targeted transcriptional control. The most popular CRISPR system comprises two essential components: a guide RNA and a multi-domain protein with Cas9 being the most commonly used.

Guide RNAs for in vitro assays are usually synthesized under the control of T7, T3 or SP6 phage promoters. Those methods require specific primers synthesized for each guide RNA, and are not suitable for large scale applications. Additionally, efficient transcription initiation starts preferably with "G" nucleotide, which poses a constrain on the first nucleotide of target sequence in the guide RNA if efficiency is a concern.
Promoters transcribed by RNA polymerase II are not suitable for controlling guide RNA in vivo synthesis because the 5' CAP, possible 5'/3' UTR and the poly A tail may hinder guide RNA function. Additionally, RNAs transcribed by RNA polymerase II are rapidly exported from the nuclei into the cytosol while nuclear localization is required for the CAS9/gRNA duplex to access the genomic DNA. Consequently, guide RNAs for in vivo assays are usually expressed under promoters transcribed by RNA polymerase III. SNR52 snoRNA promoter is commonly used in yeast, while U3/U6 snoRNA promoters are used in plants and animals. Although spatial and temporal controls of guide RNA expression is much desired in many scenarios, it could not be achieved with those promoters due to their constitutive nature.