Mechanism of coupling mRNA localization to translational activation in Drosophila
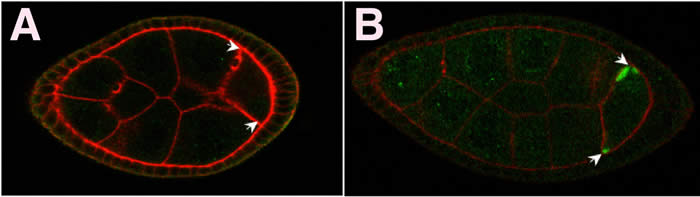
While progress has been made in identifying localization factors and translational regulators of oskar, the mechanism for coupling these processes has remained obscure. The cup gene was originally identified as a female sterile mutation that causes an open eggshell phenotype, but had no known function. Our biochemical analysis revealed that Cup binds directly to eukaryotic initiation factor 4E (eIF4E), the 5' cap binding component of the translation initiation complex. With this biochemical insight as a guide, we found that cup is a translational repressor of oskar mRNA. Furthermore, cup is also required for oskar mRNA localization and is necessary to recruit a protein essential for the plus-end directed microtubule transport of oskar. Thus, Cup is a translational repressor of oskar that is required to assemble the oskar mRNA localization machinery (Wilhelm, et al. 2003). This work suggested a general mechanism for coupling mRNA localization with translational control via common factors and demonstrated the ability of biochemical insights to lead to previously undescribed phenotypes.

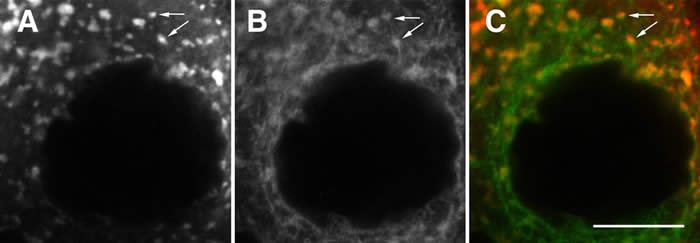
While most of the focus in the field of mRNA localization and translational control has been on regulation of key developmental molecules, it has been unclear how broad a role these two regulatory mechanisms play in basic cellular processes. Trailer hitch (tral) was identified as part of the Exu complex, however, genetic analysis revealed that tral is required for efficent secretion. We was able to reconcile these observations when we discovered that the Exu complex is present on subdomains of the endoplasmic reticulum (ER) that border ER exit sites - the sites where ER to Golgi trafficking takes place. Furthermore, tral is required for normal ER exit site formation and is associated with the mRNAs for ER exit site components. These findings have raised exciting new possibilities for how the mRNA localization machinery could interface with the classical secretory pathway to promote efficient protein trafficking in the cell (Wilhelm, et al., 2005).